Rebin with time bins aligned with orbit
from wtlike.config import *
from wtlike.cell_data import *
from wtlike.bayesian import *
from wtlike.lightcurve import *
from light_curves.b1259 import *
from utilities.ipynb_docgen import *
config = Config(data_folder='/home/burnett/weekly', verbose=1)
class B1259phased(B1259Periastron):
def __init__(self, config, first=54683, last=59326, nbins=1000, clear=False):
# construct bins with respect to phase
self.interval = interval=nbins/self.period
self.nbins = nbins
def mjd2phase_bin(mjd):
return (mjd-self.tp+self.period)*interval
def phase_bin2mjd(pb):
return pb/interval+self.tp-self.period
first_edge = int(mjd2phase_bin(first))+1
last_edge = int(mjd2phase_bin(last))
N = last_edge-first_edge
phasebins = np.array(list(map(phase_bin2mjd, np.linspace(first_edge, last_edge, N+1) )) )
super().__init__(config, bins=phasebins, bin_key='B1259_phase_bin_edges', clear=clear)
def mjd2phase(self, t):
"""return the phase, [-0.5,0.5) for t in MJD """
r = np.mod(t-self.tp, self.period)/self.period
if r>0.5: r-=1.0
return r
def combine_phases(self):
""" Fold, and fit cells to a lightcurve with respect to phase
"""
cells = self.cells
cell_width = cells.iloc[0].tw
cells.loc[:,'phase'] = cells.t.apply(lambda t: self.mjd2phase(t).round(4))
# check how many of each phase
if self.config.verbose>0:
t, c = np.unique(cells.phase, return_counts=True)
n, c = np.unique(c, return_counts=True)
fdf = pd.DataFrame(dict(cells=n, count=c))
print(f'Combine to {len(t)} phase cells: check number of cells \n {fdf}')
# group by phase
gcells = cells.groupby('phase')
phased_cells = []
for phase, gcell in gcells:
newcell=dict(t=phase*self.period, tw=cell_width)
for col in ' n e S B'.split():
newcell[col] = gcell[col].sum()
newcell['w'] = np.concatenate(list(gcell.w.values))
phased_cells.append(newcell)
self.folded_cells = df = pd.DataFrame(phased_cells)
# fold the phases, and make a folded lightCurve
self.folded_lc = LightCurve(self.config, self.folded_cells, self.source)
def do_folded_bb(self):
"""Perform Bayesian Block analysis on the folded
"""
# do the BB to get partition
self.bb_folded_edges = get_bb_partition(self.config, self.folded_lc)
# combine the cells
self.bb_folded_cells = partition_cells(self.config, self.folded_cells, self.bb_folded_edges)
# fit them
self.bb_folded_lc = fit_cells(self.config, self.bb_folded_cells,)
phase_bba=None
def B1259_phased():
"""
## Run Bayesian Block analysis wwith phased bins
Define {nbins} bins with {width:.2f}-day width.
{printout1}
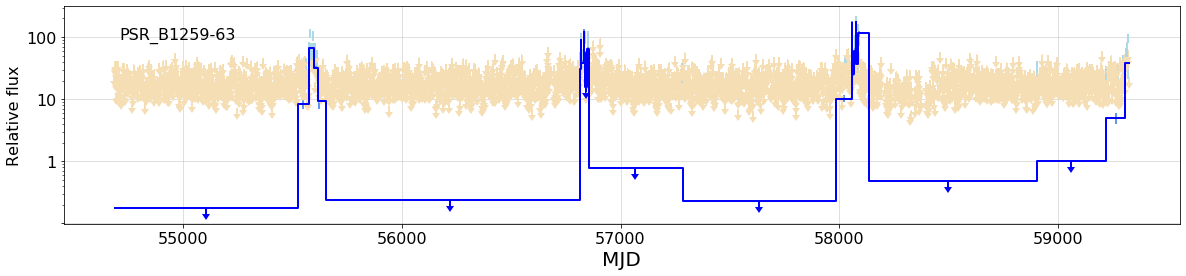
### The full light curve with BB overlay:
{fig1}
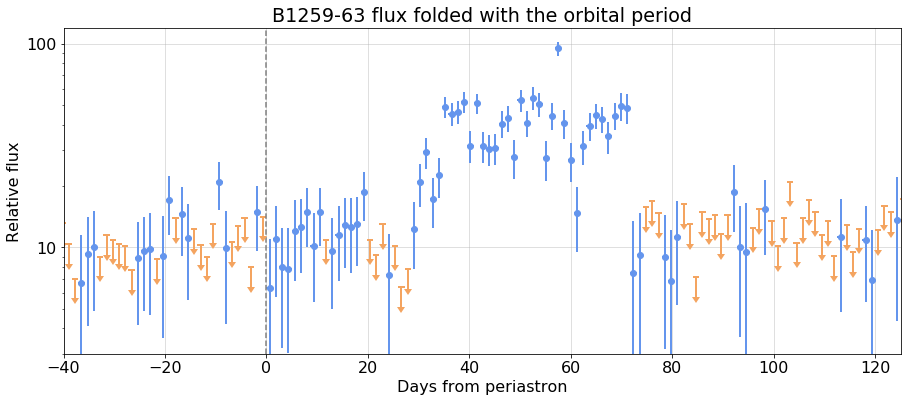
### After folding:
{fig2}
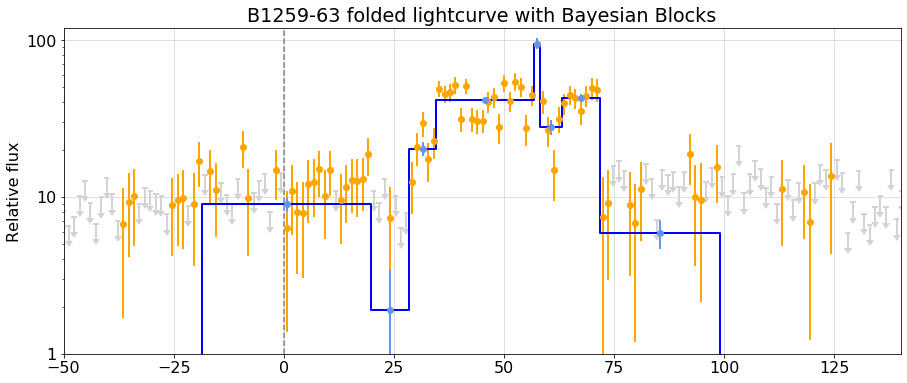
### Perform the Bayesian Block analysis on the folded light curve
{printout2}
Table of blocks, fits:
{folded_bb_table}
{fig4}
"""
plt.rc('font', size=16)
global phase_bba
with capture_print('Output from phased bin analysis') as printout1:
phase_bba = B1259phased(config)
nbins, width=phase_bba.nbins, 1/phase_bba.interval ;
phase_bba.combine_phases()
fig1 = figure(phase_bba.full_plot(fignum=1), width=800)
fig2, ax = plt.subplots(figsize=(15,6), num=2)
fig2.width=800
ax.axvline(0, color='grey', ls='--')
phase_bba.folded_lc.plot(ax=ax, xlim=(-40,125), ylim=(3,120), yscale='log', ts_min=1.5,
xlabel='Days from periastron', ylabel='Relative flux', fmt='o',
title='B1259-63 flux folded with the orbital period',
)
with capture_print('Output from Bayesian Block analysis of folded cells') as printout2:
phase_bba.do_folded_bb()
folded_lc = phase_bba.folded_lc
bb_folded_lc = phase_bba.bb_folded_lc
bb_folded_lc.loc[:,'TS'] = bb_folded_lc.fit.apply(lambda f: round(f.ts,1))
bb_folded_lc['t tw TS fit'.split()]
folded_bb_table = monospace(bb_folded_lc['t tw TS fit'.split()])
fig4, ax = plt.subplots(figsize=(15,6), num=4)
ax.axvline(0, color='grey', ls='--')
fig4.width=800
bb_overplot(config, folded_lc, bb_folded_lc, ax=ax, ts_min=1.5, xlim=(-50,140), fmt='o',
colors=' orange lightgrey blue'.split(),
yscale='log', ylim=(1,120),
title='B1259-63 folded lightcurve with Bayesian Blocks')
return locals()
nbdoc(B1259_phased)
phase_bba.weight_histogram()[:50]